The 2nd Junior Scientist Symposium will take place on September 27th from 16:00 to 17:00 (Swiss time)
Speakers:
- Hector Hernandez - Jenal Group, Biozentrum UNIBAS (Switzerland)
Title: Deciphering Pseudomonas single cell responses to metabolic and antibiotic stresses by quantitative time-lapse microscopy
Metabolic heterogeneity of bacterial populations was recently associated with their increased tolerance to antibiotics. We study the potential role of metabolic heterogeneity in antibiotic tolerance of the human pathogen Pseudomonas aeruginosa. Potential heterogeneity in central carbon utilization is reflected by the presence of three paralogous glyceraldehyde 3-phosphate dehydrogenases mediating a central step in carbon metabolism (GAPDHs: GapA, B, and C). While GapA and GapB have been associated with glycolytic fluxes, GapC mediates the reverse reaction during gluconeogenesis. We observed that in stationary phase, P. aeruginosa shows highly heterogeneous expression of gap genes, indicating that upon starvation this pathogen establishes differential metabolic programs, which cannot be assessed by population-based assays. To investigate P. aeruginosa gap gene expression at the single-cell level we used mother machine microfluidic devices, gapdhs fluorescent reporters, and quantitative single-cell time-lapse microscopy. We find that gapA and gapB are primarily expressed under glycolytic conditions (mainly during early stationary and exponential phase, respectively), whereas gapC is expressed primarily in the presence of gluconeogenic substrates, with some baseline expression under glycolytic conditions. Upon shifting carbon sources, P. aeruginosa adapts gap gene expression within the course of two hours together with reductive cell divisions and decreased growth rate. This suggests that homogenous populations are not well-equipped to cope with changing nutritional conditions and that increasing gap expression heterogeneity could represent a bet-hedging strategy to avoid non-productive lag phases after media shifts. We are currently using these new insights to investigate the role of gap gene expression heterogeneity on drug tolerance of P. aeruginosa cultures undergoing media shifts.
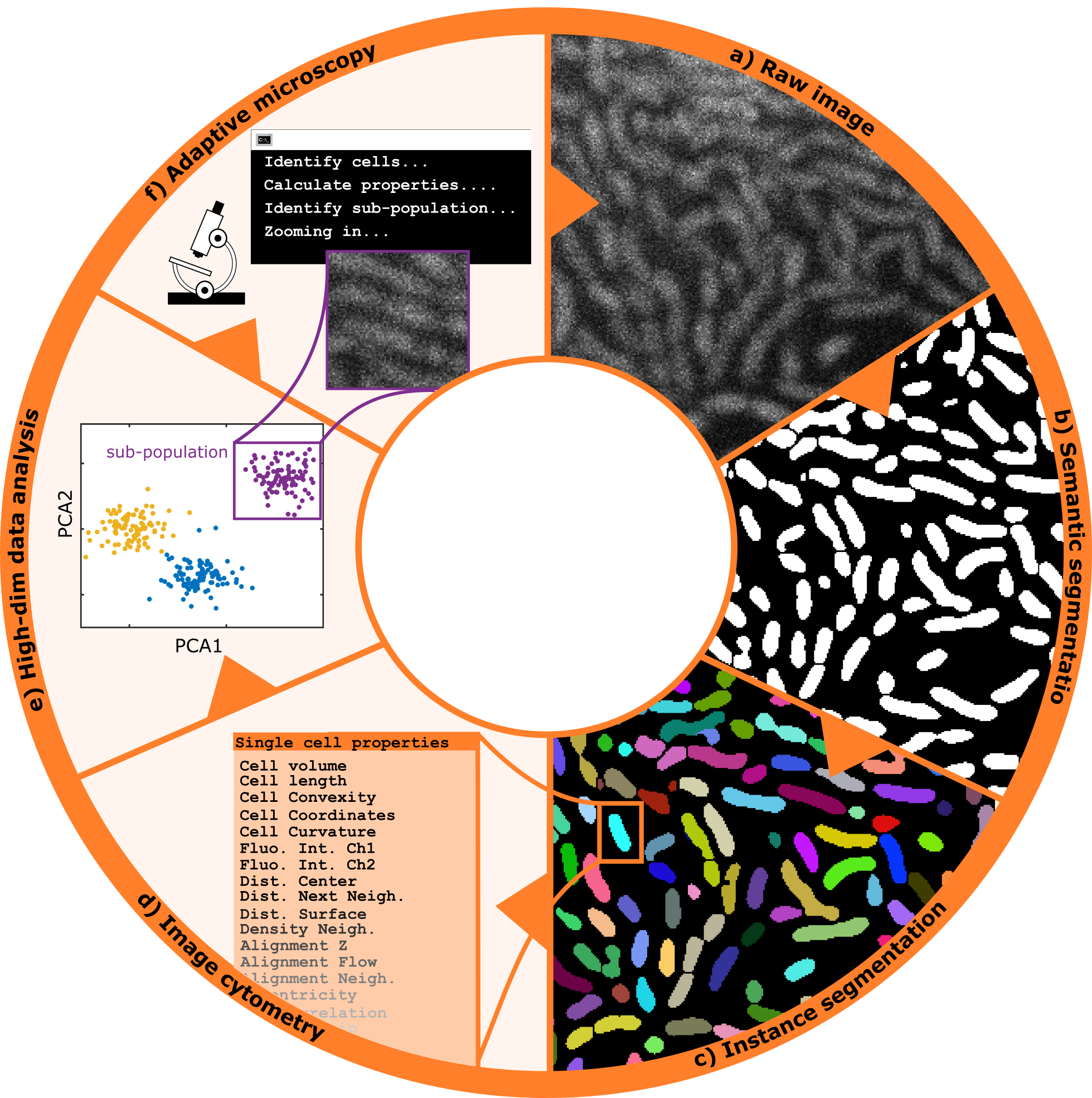
- Hannah Jeckel - Drescher Group, Biozentrum UNIBAS (Switzerland)
Title: From smart microscopy to biological insight
The introduction of novel, rapid methods of microscopy have enabled researchers to capture large amounts of data in a short amount of time, creating the necessity for approaches to automatically identify and filter for relevant information. This is traditionally done through downstream analysis after an experiment is complete, however an alternative approach is to adaptively refine the parameters of data acquisition throughout the course of the experiment, such that only relevant data is captured in the first place. In our lab, we combine image analysis techniques with microscopy control tools to create a fully automated microscope setup that adaptively performs experiments without the need for manual intervention. The data acquired through this intelligent microscopy approach is then further processed to yield a multidimensional quantification representing the relevant properties and dynamics visible in each image. Finally, dimensionality reduction and clustering techniques or similar methods can be used to obtain a distinction into subpopulations or phenotypes, which serve as the basis for an interpretation. Here, I will describe how we integrate these automated data acquisition and analysis steps into a pipeline to obtain reliable and reproducible results leading to insight into microbial systems, using bacterial swarms as an example.
- Adam Carte - Schier Group, Biozentrum UNIBAS (Switzerland)
Title: A Quantitative Microscopy Approach to Probing Nodal Activity Gradient Interpretation
Nodal signaling plays an early and essential role in the patterning of all vertebrate embryos. In zebrafish, it forms a stereotyped gradient of signaling beginning about 4 hours after fertilization that is vital to proper mesoderm and endoderm formation. Despite Nodal’s key role in development and rich history of study as a model morphogen, it remains unclear how Nodal target genes achieve differential patterns of gene expression in response to the same activity gradient. Models of Nodal interpretation suggest specific predictions about features of target gene expression in wild-type and perturbed embryos, but testing these predictions has remained elusive in the absence of necessary quantitative experimental and computational methods. Here, I present my efforts in building a toolbox of quantitative techniques to decipher how target genes interpret the Nodal gradient, highlight data that I have gathered to test specific predictions of different models of interpretation, and describe ongoing analyses designed to reveal which whether each model plays a role in generating the unique expression patterns of selected Nodal targets.
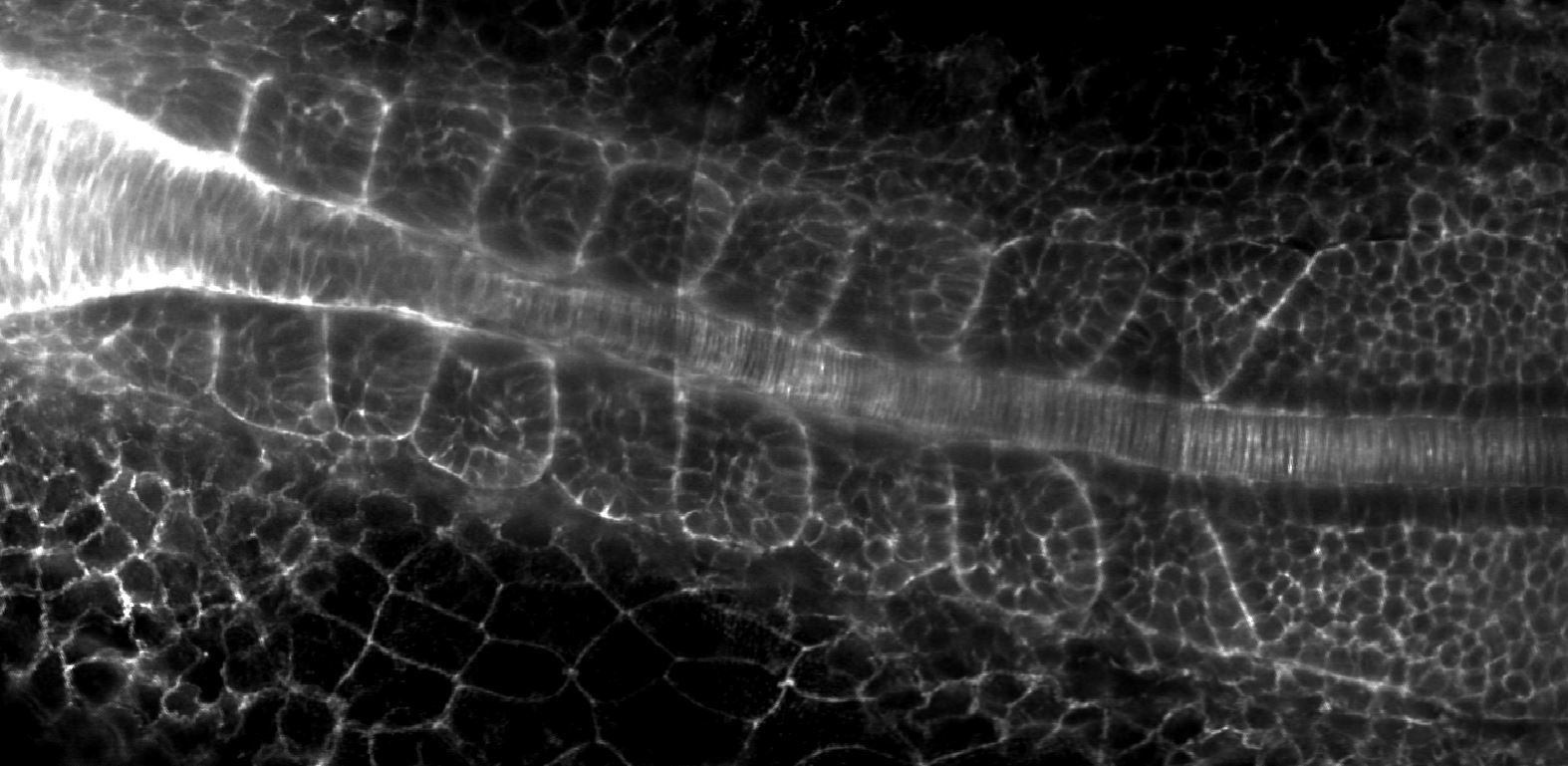
- Sundar Naganathan - TIFR Mumbai (India)
Title: Using light-sheet microscopy to study large-scale body symmetries in a developing embryo
Vertebrates are characterized by a left-right symmetric muscle and skeletal system that emerges from bilateral somites during embryonic development. Left-right symmetry is vital for adult mechanical movements and a loss of symmetry is associated with debilitating skeletal disorders such as scoliosis. Symmetry is often assumed to be a default state in somite formation, however, it remains unknown how robust somite shapes and sizes at the same position along the body axis emerge on the left and right sides of the embryo.
Towards investigating this problem, we used light-sheet microscopy to image left-right somite formation with high spatiotemporal resolution for long periods of development. By developing automated image processing tools to analyze the resulting large data sets, we could show that initial somite shapes and sizes are often asymmetric. This was in contrast to the textbook view that has often suggested that symmetry is a default state in embryonic development. Strikingly, these imprecisions are not left unchecked and we find that lengths adjust within an hour after somite formation, thereby increasing morphological symmetry. We discover an error correction mechanism, where length adjustment is facilitated by somite surface tension, which we show by comparing in vivo experiments and in vitro single-somite explant cultures with a mechanical model. We propose that tissue surface tension provides a general mechanism to adjust shapes and ensure precision and symmetry of tissues in developing embryos.