The 10th Microscopy Webinar will take place on June 28th from 16:00 to 17:00 (Swiss time)
Speaker:
Ko Sugawara - Universite de Lyon (France)
Title: ELEPHANT: cell tracking platform empowered by incremental deep learning using sparse ellipsoid annotations
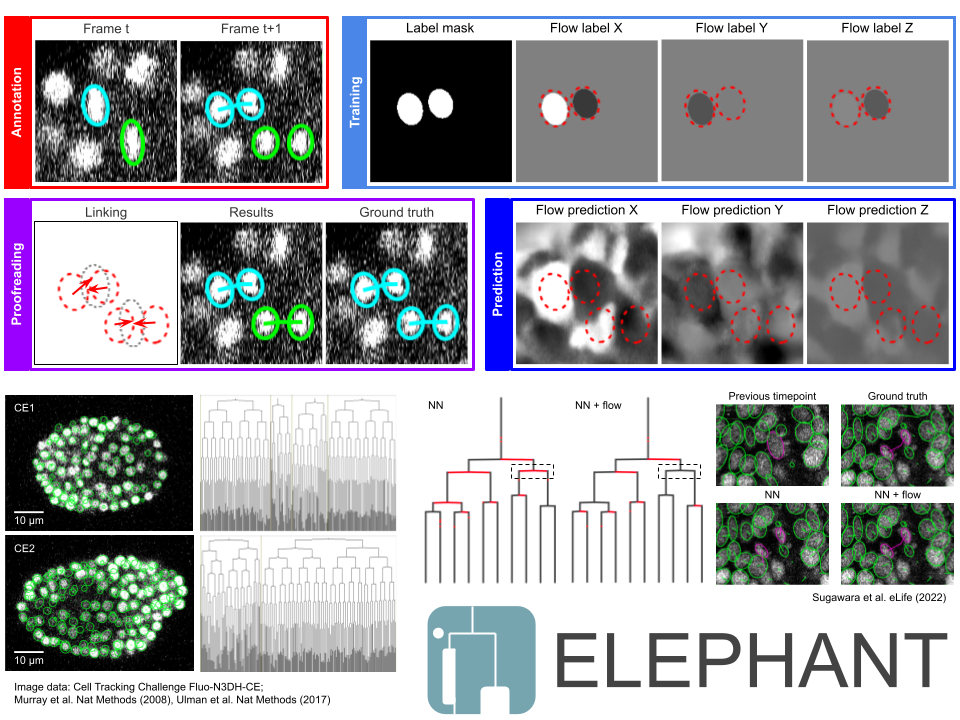
Abstract: With the advent of the deep learning era, cell tracking has become more accurate than conventional approaches. However, the wider use of deep learning in cell tracking remains limited mainly for two reasons: (i) the scarcity of annotated data, and (ii) the lack of a user interface that integrates deep learning and tracking platforms. To address these challenges, we developed ELEPHANT1, an interactive platform for cell tracking built on top of Mastodon2. The convolutional neural networks (CNNs) in ELEPHANT are designed to detect cells and to estimate their movement for supporting cell tracking. In both steps, the CNNs are incrementally trained using sparse ellipsoid annotations. Starting with just a few annotations, cycles of annotation, training, prediction, and proofreading can be efficiently iterated to enrich the tracking results. We applied ELEPHANT to the analysis of confocal microscopy data that recorded the entire process of leg regeneration in the crustacean Parhyale hawaiensis. ELEPHANT is an open source project and is available in Fiji3.
Source code: https://github.com/elephant-track
User manual: https://elephant-track.github.io
1. Sugawara, K., Cevrim C. & Averof. M. (2022) Tracking cell lineages in 3D by incremental deep
learning. eLife. doi:10.7554/eLife.69380
2. https://github.com/mastodon-sc/mastodon
3. Schindelin, J. et al. (2012). Fiji: an open-source platform for biological-image analysis. Nature
Methods, 9(7), 676–682. doi:10.1038/nmeth.2019